About
|
CITATION Kuan-Chieh Tseng, Yi-Fan Chiang-Hsieh, Hsuan Pai, Nai-Yun Wu, Han-Qin Zheng, Chi-Nga Chow, Tzong-Yi Lee, Song-Bin Chang, Na-Sheng Lin, Wen-Chi Chang, sRIS: A Small RNA Illustration System for Plant Next-Generation Sequencing Data Analysis, Plant and Cell Physiology. 2020. Background Small RNA (sRNA) is shorter than 200 nt and containg different types. Some of them like microRNA (miRNA) and short interfering RNA (siRNA) are well-known for inducing degradation of messenger RNA (mRNA). Based on this mechanism, sRNA is an important regulator for controlling gene expression. Recently, next-generation sequencing (NGS) is broadly applied to reveal regulatory pathways of sRNA in many researches. Numerous bioinformatics tools have been developed to analyze sRNA NGS data. Most of the tools focus on investigating the expression profiling of sRNA, but limit in further functional analysis. Though few of them provide some functional assay, it is hard to get useful information from their analysis outputs. Methods and Results In order to solve the problems mentioned above, a comprehensive and user-friendly system for small RNA next-generation sequencing data analysis was developed in this study. There are two packages in this system. One of the packages is for sRNA overview analysis, not only miRNA but also virus-derived small interfering RNA (vsiRNA) can be identified. The other one is for sRNA target prediction. The sRNA targets are predicted via theoretical calculation and experimental data such as degradome sequencing. Moreover, user-friendly graphical and tabular outputs are provided in the system. Conclusions This Small RNA Illustration System (sRIS) not only help user to comprehensively analyze NGS data of sRNA, but also provide easily interpretive outputs. With the useful information from the analysis outputs, it will be more convenient to discover the important regulatory mechanism of sRNA. | ||||||||
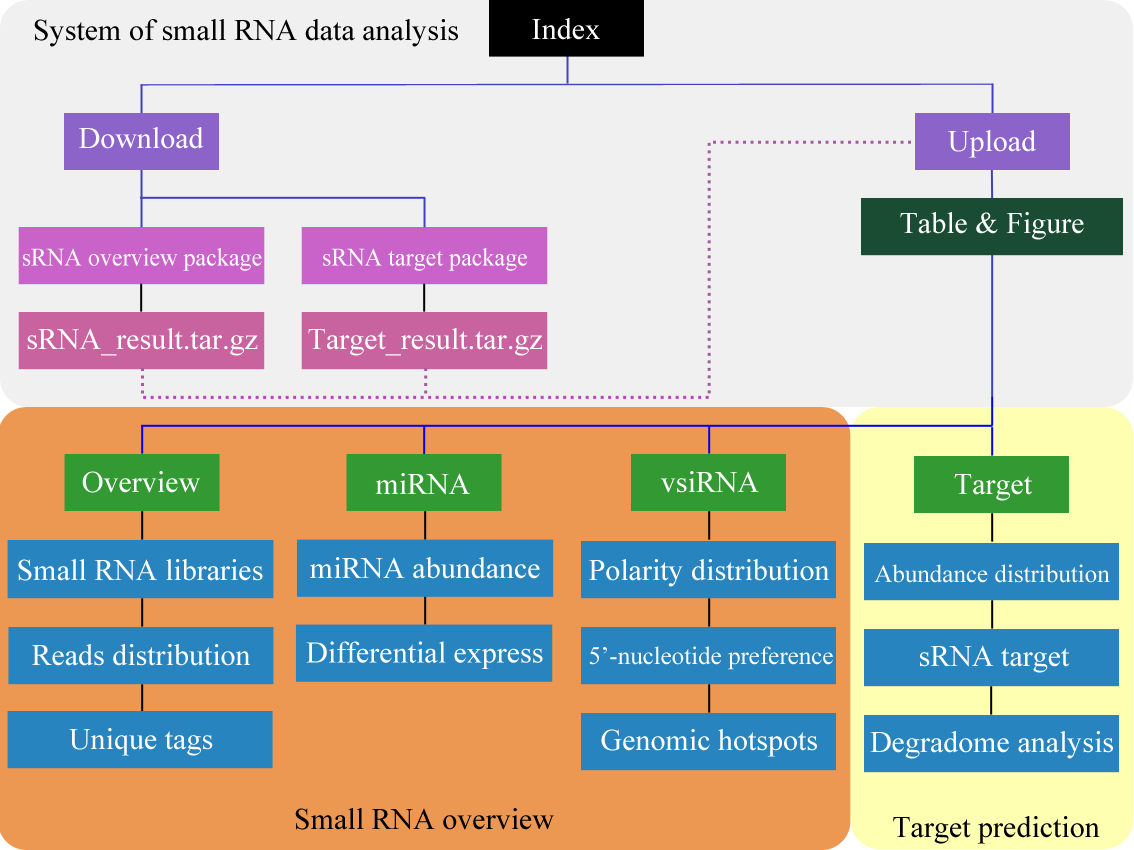 Figure 1. Construction of small RNA illustration system. | ||||||||
|
