Guide
|
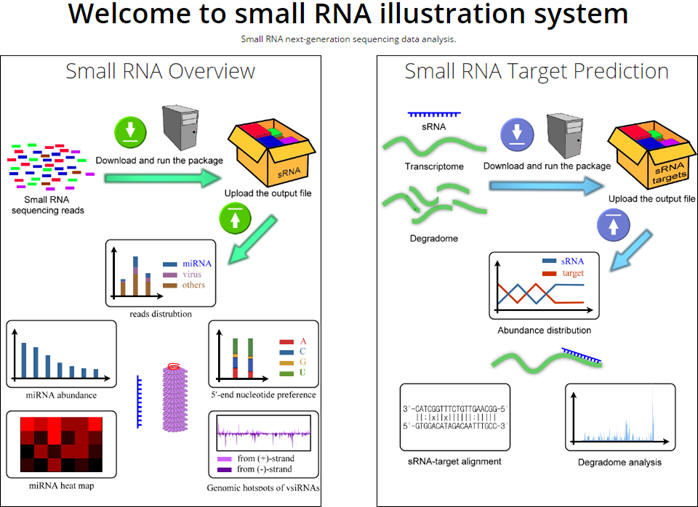
Small RNA Illustration System is a comprehensive and user-friendly system for sRNA NGS data analysis.
There are two packages providing two major analyses,small RNA overview and smll RNA target prediction. Please click on one of the figures to continue.
|
 |
|
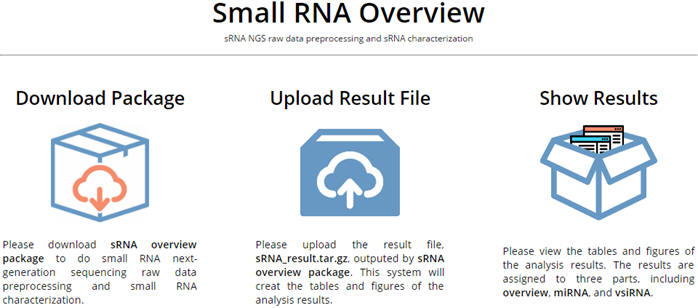
Small RNA overview is for doning sRNA characterization. To use this analysis, please follow the steps below: 1. Download sRNA overview package from the website and do the analysis of sRNA NGS data on the client side in Linux. 2. Upload the result file, sRNA_result.tar.gz, outputted by the package to the website. 3. View the figures and tables of the analysis results. |
 |
|
If it is hard for you to make command, click on "GO" for help.
|
 |
|
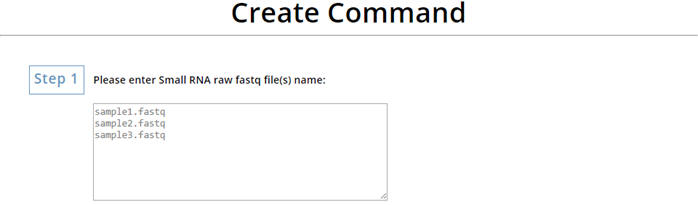
Enter the file name of your sRNA NGS raw data.
|
 |
|
Enter the read length you want to remove.
|
 |
|
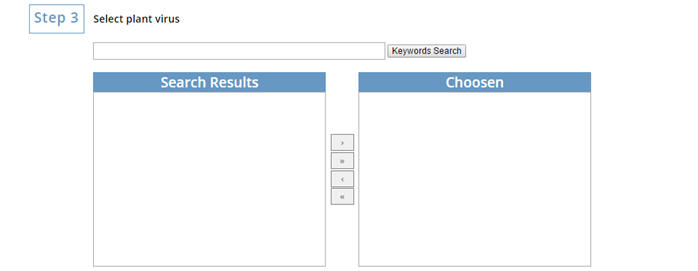
Search the plant virus you want to use.
|
 |
|
Select the EST you want to use or enter your own file name.
|
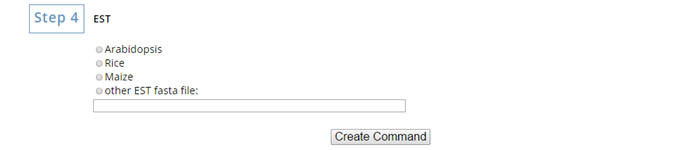 Finally, click on "Create Command" to get the command for running sRNA overview package. |
|
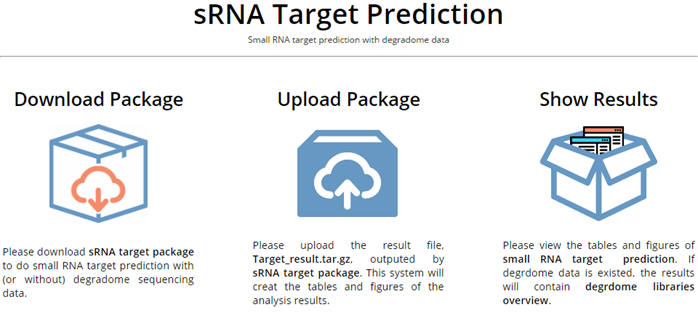
To do small RNA target prediction, please follow the steps below: 1. Download sRNA target package from the website and do the analysis of sRNA target prediction with/without degradome on the client side in Linux. 2. Upload the result file, Target_result.tar.gz, outputted by the package to the website. 3. View the figures and tables of the analysis results. |
 |
|
If it is hard for you to make command, click on one of the
buttons for help.
|
 |
|
Note:
Here use "with Degradome" as example. There are two commands for
two Perl scripts, create_sample_use.pl and . 1. Creat command for "create_sample_use.pl" Enter the file name of your Degradome raw data. |
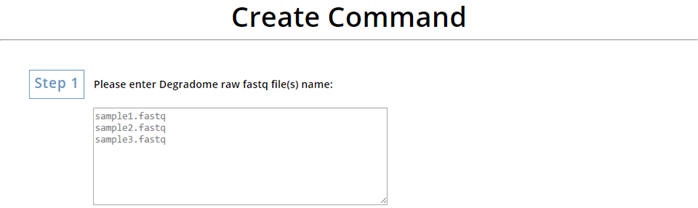 |
|
Enter the adaptor of your each degradome data. |
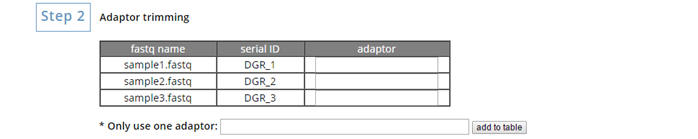 |
|
Enter the name of each sample you want to show on the figures and tables.
|
 |
|
Enter the file name of sRNA expression data.
|
 |
|
Enter the name of directory for package to output results.
|
 Finally, click on "Create Command" to get the command for running "create_sample_use.pl". |
|
2. Creat command for "run_Degradome_analysis.pl" Select the EST you want to use or enter your own file name. Note:You can create command now for running "run_Degradome_analysis.pl" or click "option" for more option. |
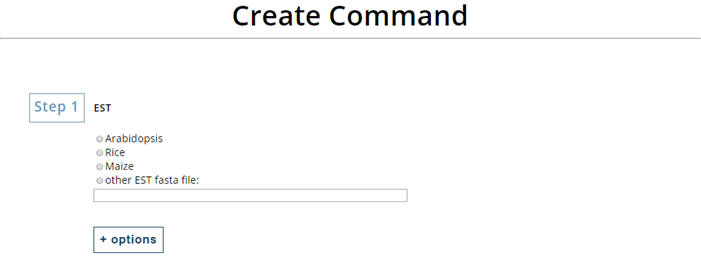 |
|
Select if you want to analyze vsiRNA.
|
 |
|
Enter the name of file conatin Rfam.
|
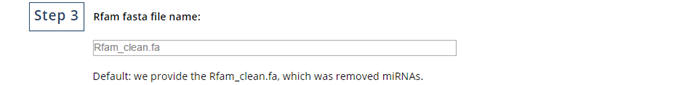 |
|
Enter the length of degradome reads you want to retain.
|
 |
|
Select if the predicted target site must between 10th and 10th base of miRNA.
|
 |
|
Enter the path of Bowtie.
|
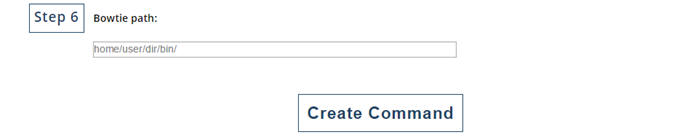 Finally, click on "Create Command" to get the command for running "run_Degradome_analysis.pl". |
